Projekt
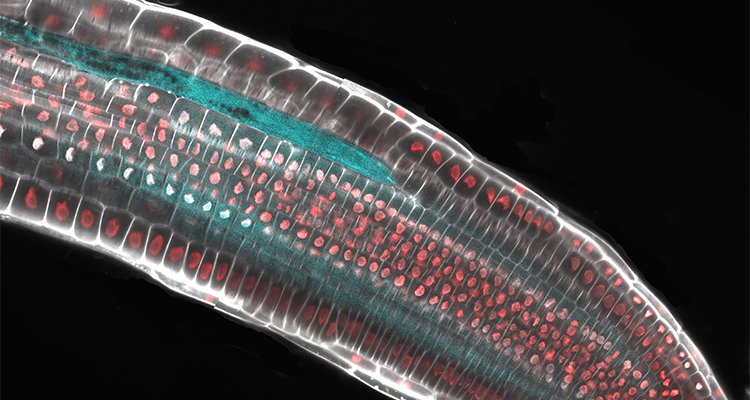
Spatial transcriptomics of roots infected with nematodes
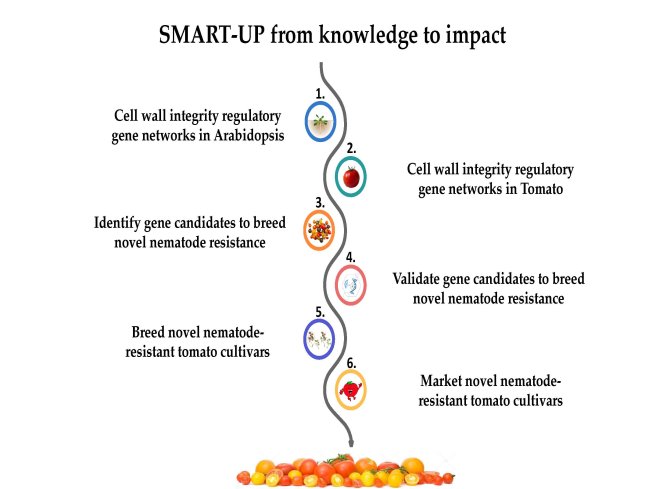
We aim to decipher the cellular and molecular mechanisms underlying cell wall integrity upon nematode parasitism, which ultimately contributes to the development of broad-spectrum resistance to nematodes in major crops.
Background
Plant parasitic nematodes cause substantial economic losses in agricultural production worldwide. Current control measurements, such as resistance genes and pesticides, are not durable or are toxic to the environment. Hence, we study the early stages of nematode infection in Arabidopsis and tomato to find a long-lasting, broad-spectrum resistance against nematodes. Knowledge of the early stages of nematode feeding site development will help the breeding industry to develop nematode-resistant crops.
Project description
We use state-of-the-art technologies to study the early stages of nematode feeding site development. Using a pioneering spatial transcriptomics technology called RNA tomography, we generate transcriptomic maps to select candidate genes involved in cell wall integrity perception during nematode infection. These candidate genes are chosen and validated using advanced bioinformatic analyses and CRISPR genome editing. Additionally, three-dimensional models of the feeding sites in plant roots are reconstructed using leading-edge microscopy and spectroscopy. A toolkit based on machine learning and deep learning will be developed to facilitate the integration of spatial transcriptomics and microscopy data in plants.