Project
Joseline Houwman - Cotranslational folding of a ribosome-bound α-β parallel protein
Proper folding of proteins is of vital importance for all living organisms. Much research has been focused on in vitro folding pathways and how misfolded intermediates arise. However, in vitro studies do not reflect the actual situation in the cell, where the protein chains are produced in a linear fashion and chaperones are abundant to assist the correct folding of these nascent polypeptides.
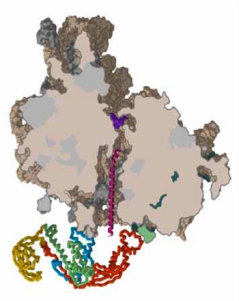
In this project we will produce ribosomal nascent chains (RNC’s), which consist of a polypeptide chain anchored by its C-terminus to the ribosome. The secondary and tertiary structure of the polypeptide will be determined by proteolysis and other structure determination techniques, such as NMR. By looking at different lengths of the polypeptide, we will obtain snapshots of the protein while it is being produced on the ribosome.
The protein used in this project is flavodoxin, which has been studied in vitro in great detail. The protein has a α-β parallel fold, which is one of the most common folds found in proteins. A mutant of flavodoxin is available which misfolds under controlled conditions and which may provide valuable insights into the rescue mechanisms of the cell.