
Project
Building preparedness challenges for viral outbreaks in the agro-food and environmental domains
The goal of the project is to build the infrastructure to enable early detection and rapid response to potential cross-species (animal-human) transmission events. Such infrastructure is currently underdeveloped, yet it is critical for the prevention of zoonoses. We will develop tools for: 1) prediction and identification of potential risks for spill over events leading to local or epidemic transmission and 2) real-time monitoring and response to outbreaks.
The project comprises cohesive projects (2) and work packages (8) which will each cover one aspect of preparedness for emerging zoonotic viruses (ZV).
Project 1. Toolbox for early prediction and analysis potential zoonotic viruses
The vast majority of ZV evolve in (wild) animals, often in circulation between multiple hosts (wildlife, livestock and humans). However, we have little insight in the vast biodiversity of viruses, or in the transmission routes of ZV between people, wastewater, urban wildlife, food and feed (Figure 1). We are limited in our ability to predict whether new mutations can be potentially dangerous for livestock and humans.
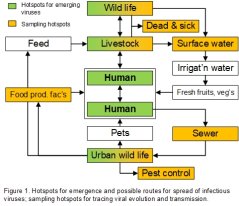
The aim of project 1 is to gain a better understanding of the origin, evolution and transmission routes of a number of virus families known to cause respiratory and gastro-intestinal infections, and whose appearance and spread is intimately associated with the food chain. Our aim is to develop metagenomics tools that detect changes in the genetic make-up of ZV as early as possible. The project proposes to deliver the following tools:
- WP 1.1. Genomic tools
- WP 1.2. AI models to predict virus outbreaks
- WP 1.3. Centralized big data infrastructure
Project 2. Toolbox for monitoring early spread of emerging zoonotic viruses
In response to early indications that potentially zoonotic viruses circulate in wildlife species, it is important to develop sensitive diagnostics. These diagnostics need to allow 1) detection of such previously uncharacterized virus variants with associated zoonotic risks in wildlife, livestock and humans; 2) a platform for readiness to ramp up routine diagnostics in an outbreak scenario. One of the lessons of SARS-CoV-2 is that in the absence of reliable diagnostic processes, a pandemic can trigger the uncontrolled introduction of unvalidated and unreliable tests to the public. Currently it is painfully apparent that scaling up of reliable tests is far too slow, significantly hampering the ability of public health bodies to control the outbreak.

The aim of project 2 is to create a toolbox of detection formats that allow rapid-release and ramp-up of diagnostics of emerging viruses:
- WP 2.1. Bioinformatics for test development and in silico validation
- WP 2.2. On-site and lab tests utilizing CRIPSR-Cas based detection
- WP 2.3. On-site and lab tests utilizing LAMP based detection
- WP 2.4. Serological methods for early outbreak diagnostics
- WP 2.5. Molecular infectivity tests for viruses
Want to know more?
Please get in touch with Andries Koops or Wim van der Poel via the contact form below.