
Use case
Improving the utilisation of meat inspection data in pig slaughterhouses for health surveillance monitoring
At slaughter, all pigs are visually monitored for health problems. The results are reported back to the farmer, as a signal and to be used for taking on-farm measures to reduce such aberrations. This kind of slaughter line inspection generates large amounts of data. To date, utilisation of this information is limited to the farm level.
Besides production of within farm incidences and trends, further use is scarce. The analysis and interpretation of slaughter inspection data is a complex task as the health status of batches of animals depends on multiple inter-related variables, also beyond farm level, such as season, day of the week, time, systematic slaughterhouse differences and observer effects. Combined analysis of the data (across farms) is more powerful in detecting patterns, trends and deviations than analysis for farms separately. In other words: farms could profit from the information in other farms’ data. Regarding the complexity and volume of slaughter line data, a big data approach is seen as a useful alternative to conventional statistics.
Our approach: machine learning systems vs linear models
Machine Learning has become increasingly important with the aim of automatically learning from data, particularly in situations where large collections of heterogeneous data are available. This study assessed the performance of machine learning algorithms using a pre-processed and a fuzzy dataset in analysing and predicting lung health aberrations based on slaughter line inspection data. Moreover, performance of these machine learning algorithms were compared with performance of a, more traditional, statistical model. Meat inspection data were provided by two Dutch slaughterhouses and collected between January 2011 and July 2014. A cleaned data set from 119 farms, comprising 1,694,158 pigs slaughtered in 12,419 batches and a less structured dataset comprising of 2,292 farms and 5,770,310 pigs slaughtered in 47,819 batches were used for analyses. Linear model (full linear model) and machine learning methods (RandomForest, ExtremeLearning, GradientBoosting, NeuralNetworks) were compared in predicting batch incidence of pneumonia. Parameters available comprised information on farm, slaughterhouse, period (week, season etc.), batch size. Moreover, to take advantage of historical information, six pneumonia-derived variables were extracted from the raw data: pneumonia percentage previous batch, rolling mean, rolling median, rolling standard deviation, rolling coefficient of variance and rolling autocorrelation of the five previous batches of a farm.
(Expected) impact of the approach
Across farm analysis substantially enhanced quality of pneumonia monitoring by reducing between batch variation by 20-30%, which reduces the uncertainty of the farm information. Both in the cleaned and in the less structured data set, machine learning was superior to classic linear statistics. Especially the RandomForest method produced better results: prediction errors were on average 10% lower, which further enhances the diagnostic and predictive quality of the farm figures. A more elaborate analysis of the results (prediction errors) revealed that the superiority of machine learning was not consistent over categories of farms. Its benefits over conventional statistics in farms with medium to high pneumonia incidences was counteracted by its failure to make good predictions for farms with less health problems. This observation supports the view that a big data analysis requires an informed (human) eye, and cannot be used as ‘automated analysis’.
Facts & Figures
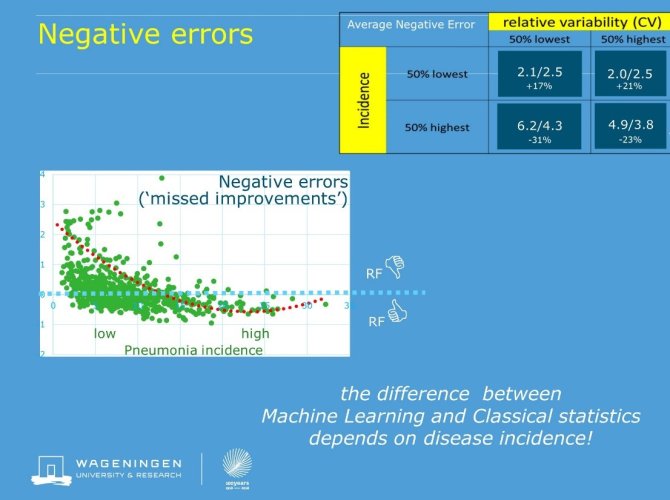
Figure: negative errors (overestimations of Pneumonia incidence) of RandomForest (RF) relative to conventional statistics. Values above 0 imply worse performance of RF
Tools used
- RandomForest
- ExtremeLearning
- GradientBoosting
- NeuralNetworks
- Using the CARET package in R
In cooperation with
- VION