
Project
Bonds resisting stress by becoming stronger
Catch bonds are protein interactions that strengthen under force. They are important for the perception of mechanical stimuli in mammalian cells. However, no examples have been found in walled cells (plants, fungi, oomycetes). There is a hyperfocus on mammalian cells, because current methods that test whether proteins can form catch bonds are incredibly time consuming. Our aim is to identify catch bonds from walled organisms by using a new microfluidic screening technique, and to study their biological relevance and function.
Background
Catch bonds have currently been identified in mammalian cells and bacteria, where they play a crucial role in mechano-biological functions. For example, the bacteria that cause a urinary tract infection use catch-bonds to adhere to the surface of our bladder, and when the catch-bond function of a protein bundling actin filaments together is lost, mechanical tissue failure ensues.
There are many more catch-bonds waiting to be discovered, and it stands to reason that they are present in all kingdoms of life. In this project we will search and study protein catch bonds from walled cells, such as plants and their pathogens. Plants, for example, cannot move in response to a threat, yet they can quickly and locally respond, for example when a pathogen tries to invade its cells. Plants accomplish this fast response without having a central nervous system that can identify the existence and location of the threat. How plants perceive and respond to these mechanical forces remains unknown. Proteinaceous catch bonds could be important elements in the way a plant deals with mechanical stimuli from the environment.
Project description
The aim of this project is to identify new catch-bonds from walled organisms and study their function from a biophysical-chemistry perspective.
The mechanical response of purified proteins will be studied using microfluidic force spectroscopy. As the name suggests, the method uses a laminar flow field in a microfluidic device to impose well-defined mechanical forces on the interaction (i.e. “bond”) of interest. The molecules of interest, e.g. potential catch-bonds, are attached to micrometer-sized beads. The beads interact with the surface of the microfluidic device, which is functionalised to carry the interaction partner. By analysing the motion of the beads, e.g. bead rolling or detachment, the mechanical response of the interaction can be inferred. A schematic overview of the method is shown below.
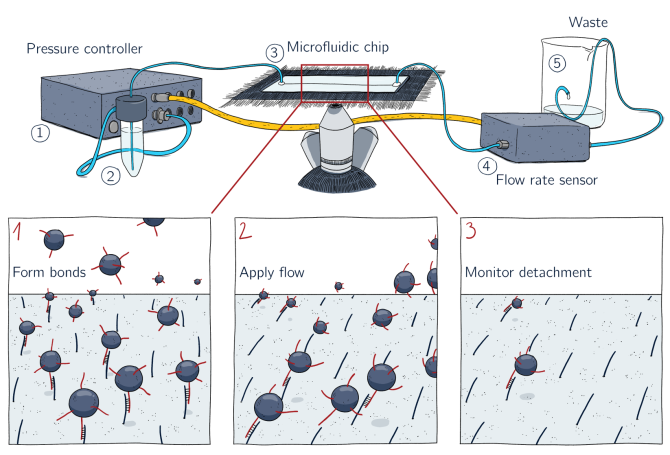
Our current focus is on catch bonds that form part of the actin cytoskeleton, which is a structure that is known to respond rapidly, locally and quantitatively to mechanical stress. In addition to molecular mechanotyping, we will use quantitative confocal fluorescence imaging in reconstituted cytoskeletal networks, formed in-vitro from purified components, for example combined with optical tweezing, to study how these putative catch bonds shape the mechanics of cytoskeletal networks. An example of the in vitro cytoskeletal network is shown in the leading figure, which I acquired at the EmbioSysLab of PCC. To create a full story, the mechanical responses of the protein catch bonds are combined with biochemical analysis, for example the protein structure and the binding affinity.
New candidate catch bonds in the plant and pathogen cytoskeletal proteome also need to be found, which is done in collaboration with PhD student Kasper Arfman. He uses artificial intelligence models for protein structure to identify homologs of known catch bonds in the animal kingdom in plants and the pathogen Phytophthora.
Additionally, we will explore how these catch bonds function in their biological context by studying the relevance of the catch bond behaviour for mechanoperception in-vivo. This is done in collaboration with postdoc Jochem Bronkhorst.
Contact
Do you have a question about plants and mechanical stimulation, or would you like to join us as a student researcher? Please contact us.