Project
Improving photosynthesis by understanding cyto-nuclear interactions
Over one billion years ago the eukaryotic plant cell originated from the endosymbiosis of an early eukaryotic cell and a photosynthesizing cyanobacterium, resulting in eukaryotic cells containing plastids and mitochondria as cytoplasmic organelles still found in all present-day plants.
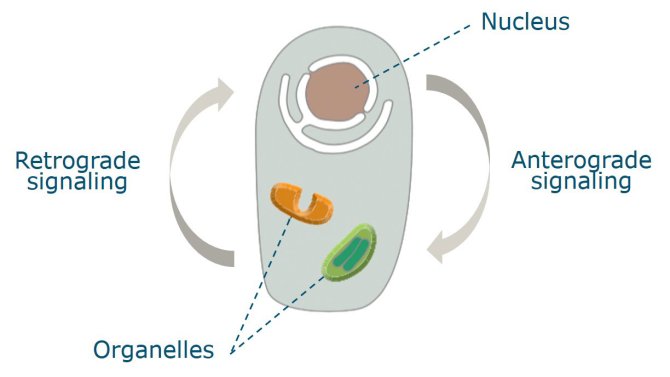
In the course of evolution, the genomes of these organelles reduced in size and gene load and they currently encode only a small fraction of the proteins needed for their functioning. As a result the symbionts have progressively fallen under the suzerainty of the host. This loss of autonomy on the part of the symbionts is not, however, complete. The remaining organellar genes are essential and they interact with nuclear genes to ensure proper plant cell functioning, involving anterograde and retrograde signalling (Figure 1). The degree to which variation in the nuclear and cytoplasmic genomes influences this interaction is not well understood.
Traditionally, plant breeding has focused on nuclear-encoded crop traits and has hardly assessed variation in the cytoplasmic contribution to traits, or the interactions between nuclear genome and cytoplasmic genomes (the so-called cyto-nuclear interactions). In retrospect this is surprising considering that chloroplasts and mitochondria are of major importance in carbon assimilation and respiration in plant cells, processes that are fundamental to plant growth. Hence, the ability to account for cyto-nuclear interactions in the selection and breeding process could offer important new opportunities to develop plant varieties that are more nutrient use efficient and robust, and which provide stable and secure yield improvements over a range of geographical locations, climates and agricultural practices.
We use Arabidopsis thaliana to study cyto-nuclear interactions in plants. We investigate both the physiological consequences of the cyto-nuclear interactions, which affect plant performance, such as photosynthesis efficiency and growth, as well as the causal genetic factors underlying the observed phenotypic variation. A further understanding of these interactions will open new options for crop breeding, progressing beyond the marker assisted selection of nuclear encoded traits, and will provide a new perspective on the evolutionary ecology of plant adaptation to a changing environment.