Project
Biocatalytic exploitation of monooxygenases
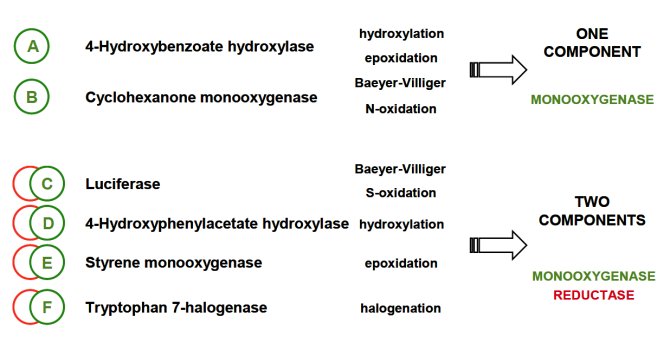
Rhodococcus strains are rich in oxidative catalysts. However, up to now only few of these enzymes have been explored for their synthetic value. Preliminary analysis has indicated that the available rhodococcal genomes harbor a large number of flavin- and heme-dependent monooxygenase encoding genes. Several of them are encoding known homologs while the majority has no clear known ortholog. In this project we apply comparative genomics to predict the protein function of putative rhodococcal monooxygenase encoding genes. Special focus is on hydroxylases and epoxidases that are active with aromatic, polycyclic and heterocyclic compounds.
Approach:
-Bioinformatic exploration of Rhodococcus genome sequences
-Genome harvesting of flavoprotein hydroxylases and epoxidases
-Biocatalytic screening of the obtained monooxygenases using a defined set of target molecules
-Redesign of interesting biocatalysts to meet the demands of a biocatalytic application