Software
fitPoly
fitPoly is an R package fro genotype calling of polyploid individuals based on bi-allelic marker assays such as Axiom, Infinium and KASP arrays. The main characteristic of such assays is that they produce two signals for a sample for a given marker, one signal for each of the two alleles.
The functions in fitPoly take as input the ratios of these two signals, and fit a mixture model to the distribution of these ratios. The mixture model has one component distributions for each of the (ploidy +1) possible allele dosages.
fit Poly is the successor of the earlier package fitTetra. It is not limited to tetraploids but can fit data for any ploidy level. In addition, it is possible to define multiple subpopulations among the samples, each with a different segregation ratio. These segregation ratios can be free, constricted to Hardy-Weinberg ratios or constructed to full-sib (FS) family ratios, if FS parents and progeny are defined.
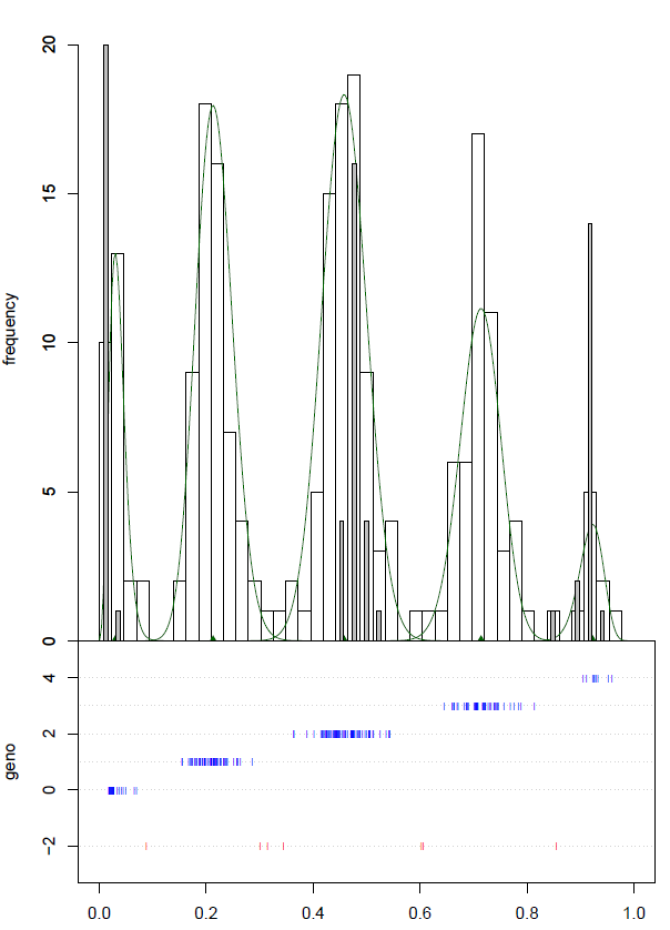
The main user function of the package, saveMarkerModels, iterates over all or a selected set of markers, fits a series of mixture models for each marker and selects the best model according to several criteria. Based on further criteria it then either rejects the marker, or it uses the selected model to assign genotype scores to each sample. Samples that cannot be genotyped reliably are assigned a missing value for the genotype. The output consists of a table with data on the fitted model per marker and a table with a.o. the assigned genotypes for each sample per marker. Optional further output includes a log file, a table with data on all fitted models per marker, and graphical output (e.g. Figure 1).