
Equipment
Confocal Laser Scanning Microscopy (CLSM)
Confocal laser scanning microscopy (CLSM) allows 3D localization of labelled target molecules in cells and food samples.
Confocal laser scanning microscopy (CLSM) allows 3D localization of labelled target molecules in cells. The principle of a confocal is that an aperture (pinhole) is placed in the image plane, so most of the out-of-focus light (orange in figure) will not reach the detector. This will not only enhance the signal to noise ratio (enhanced contrast), but will also give rise to a better sectioning (thinner slices in z-direction).
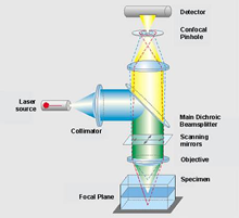
The following information can be obtained using CLSM.
- (Co)-localisation: Specific fluorescent probes (chemical dyes or fluorescent proteins technology) allow temporal and spatial colocalisation of (bio)molecules of interest. A range of fluorescent probes can be combined for simultaneous multicolor, multidimensional image acquisition. Specific applications like time-laps imaging or fluorescence recovery after photobleaching (FRAP) can be performed.
- 3D reconstruction: Optical sectioning (nm resolution) can be achieved using the piezo stage of the confocal microscope, allowing for 3D-image reconstruction of (bio)molecules, organelles and tissues. 3D-image reconstruction can also be performed using simultaneous multi-color image acquisition.
- Fluorescence Resonance Energy Transfer (FRET) by acceptor photobleaching. FRET is the energy transfer through dipole-dipole coupling of an donor and acceptor chromophore. and can be used to study protein-protein interactions. A protocol for detecting FRET by acceptor photobleaching is available. For quantitative FRET measurements, FRET-FLIM is more suitable.
- Imaging lipid and protein content in food. A protocol for staining lipid and protein particles in food samples is available. Can be used for 3D imaging of food ultrastructure with a resolution of approximately 250 nm (XY) and 1μm (Z). For most food samples, the imaging depth is 50 μm.
Both a Zeiss LSM510 (for standard measurements) and a Leica SP5 (for advanced measurements) are available.
Zeiss LSM510
The confocal laser scanning microscope (CLSM) is typically used for imaging of cells or food samples. The instrument is based on an inverted microscope with a LSM-detector head coupled to the base port of the microscope. Excitation light is provided by an argon ion laser(458, 488 and 514 nm) and two helium neon lasers (543 and 633 nm).

All laser lines are combined and connected to the scanner head by means of a fiber optic. An Acousto Optical Tunable Filter (AOTF) allows to vary the intensity of laser light and enables to select excitation light of any combination. The LSM head contains a large set of excitation, dichroic and emission filters, scanning mirrors, size adjustable pinholes and three sensitive photomultiplier tubes. Several water, oil and air immersion objectives (Zeiss) are present. This fully motorized system is PC-controlled and allows the real time monitoring of non-processed and ratio images.
Leica SP5
The Leica SP5X-SMD high-end fluorescence microscope, dedicated for imaging cells and multicellular system like plant roots. The instrument is based on an inverted microscope with a scanhead coupled to the base port of the microscope. It combines confocal imaging with single molecule detection (SMD). The SP5 has variable excitation with a supercontinuum tunable white light laser (WLL) source. WLL allows confocal imaging using up to eight excitation wavelengths simultaneously. Confocal microscopy with five detection channels including hybrid detector technology allows for complete, filter free, spectral freedom in imaging. Automatic 3D image acquisition using a third parameter like wavelength, time or volume.

The main feature of the SP5 is the FLIM option. Fluorescence lifetime imaging microscopy is done by the WLL tunable pulsed excitation and TCSPC detection (both Becker&Hickl and Picoquant systems are available). FLIM acquisition has a time resolution of 100 ps or better and short detector dead time in combination with a high dynamic range.
Publications
- Frei dit Frey, N., Mbengue, M., Kwaaitaal, M.A.C.J., Nitsch, L.M.C., Altenbach, D., Häweker, H., Lozano-Duran, R., Njo, M.F., Beeckman, T., Huettel, B., Borst, J.W., Panstruga, R. & Robatzek, S. (2012). Plasma membrane calcium ATPases are important components of receptor-mediated signaling in plant immune responses and development. Plant Physiology, 159(2), 798-809.
- Rademacher, E.H., Lokerse, A.S., Schlereth, A., Llavata Peris, C.I., Bayer, M., Kientz, M., Freire Rios, A., Borst, J.W., Lukowitz, W., Juergens, G. & Weijers, D. (2012). Different Auxin Response Machineries Control Distinct Cell Fates in the Early Plant Embryo. Developmental cell, 22(1), 211-222.
- De Rybel, B., Berg, W.A.M. van den, Lokerse, A.S., Liao, C.Y., Mourik, H. van, Moller, B.K., Llavata Peris, C.I. & Weijers, D. (2011). A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiology, 156, 1292-1299.
- Esse, G.W. van, Westphal, A.H., SURENDRAN, R.P., Albrecht, C., Veen, M.B. van, Borst, J.W. & Vries, S.C. de (2011). Quantification of the BRI1 receptor in planta. Plant Physiology, 156(4), 1691-1700.
- Rademacher, E.H., Moller, B.K., Lokerse, A.S., Llavata Peris, C.I., Berg, W.A.M. van den & Weijers, D. (2011). A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. The Plant Journal, 68, 597-606.
- Slootweg, E.J., Roosien, J., Spiridon, L.N., Petrescu, A.J., Tameling, W.I.L., Joosten, M.H.A.J., Pomp, H., Schaik, C.C. van, Dees, R.H.L., Borst, J.W., Smant, G., Schots, A., Bakker, J. & Goverse, A. (2010). Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. The Plant Cell, 22(12), 4195-4215.
- Viotti, C., Bubeck, J., Stierhof, Y.D., Krebs, M., Langhans, M., Berg, W.A.M. van den, Dongen, W.M.A.M. van, Richter, S., Geldner, N., Takano, J., Juergens, G., Vries, S.C. de, Robinson, D.G. & Schumacher, K. (2010). Endocytic and Secretory Traffic in Arabidopsis Merge in the Trans-Golgi Network/Early Endosome, an Independent and Highly Dynamic Organelle. The Plant Cell, 22(4), 1344-1357.
- Immink, G.H., Tonaco, I.A.N., Folter, S. de, Shchennikova, A., Dijk, A.D.J., Busscher-Lange, J., Borst, J.W. & Angenent, G.C. (2009). SEPALLATA3: the 'glue' for MADS box transcription factor complex formation. Genome Biology, 10, R24.
- Rademacher, E.H. & Weijers, D. (2007). Got root? Initiation of the embryonic root meristem. International Journal of Plant Developmental Biology, 1(1), 122-126.
- Bosgraaf, L., Waijer, A., Engel, R., Visser, A.J.W.G., Wessels, D., Soll, D. & Haastert, P.J.M. van (2005). RasGEF-containing proteins GbpC and GbpD have different effects on cell polarity and chemotaxis in Dictyostelium. Journal of Cell Science, 118(9), 1899-1910.
- Pouwels, J., Velden, T. van der, Willemse, J., Borst, J.W., Lent, J.W.M. van, Bisseling, T. & Wellink, J.E. (2004). Studies on the origin and structure of tubules made by the movement protein of Cowpea mosaic virus. Journal of General Virology, 85(12), 3787-3796.
- Burg, H.A. van den, Westerink, N., Francoijs, C.J.J., Roth, R., Woestenenk, E.A., Boeren, J.A., Wit, P.J.G.M. de, Joosten, M.H.A.J. & Vervoort, J.J.M. (2003). Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. Journal of Biological Chemistry, 278(30), 27340-27346.