Project
Quantifying Electron Fluxes In Methanogenic Microbial Communities
Anaerobic digestion of organic residues is a classical example of a process that converts waste into a renewable resource, i.e. methane containing biogas. Unlike the production of other biofuels, biogas production from waste is not associated with negative aspects like competition with food production or the utilization of enormous surfaces of arable land. Methanogenesis is a naturally occurring process that takes place in anaerobic environments in which organic matter is degraded. Methanogenesis under controlled conditions in bioreactors is a sustainable way to conserve chemical energy in organic residues as biogas. Since methane is the organic compound with the lowest energy content per electron, all kind of organic waste components are funnelled to biogas when degraded anaerobically.
Despite the fact that anaerobic digestion is widely applied on a large scale, remarkably little quantitative understanding is available about the exchange of metabolites between the different microbial groups involved in the conversion of complex substrates to biogas. Therefore the anaerobic digestion process is typically treated as a black box process, and consequently only pragmatic tools are available to analyse the process.
This project aims to open the black box of anaerobic digestion by getting a detailed understanding of electron fluxes occurring in the ultimate steps of the process (methanogenesis). A more mechanistically correct process description will result in the development of possibilities for enhanced process operation, process control, and future bioreactor design. The project aims at acquiring the fundamental knowledge needed for establishment of an advanced tool for anaerobic digester system analysis.
Insight into the key features of methane formation by syntrophic communities will be obtained by analysing defined cocultures constructed from pure cultures of acetogenic bacteria and methanogens. Microorganisms of which the genome information is available will be used. This will allow to study communities at molecular, cell and community level.
The microorganisms of choice include: Syntrophobacter fumaroxidans (propionate degrader), Syntrophomonas wolfei (butyrate degrader), Methanospirillum hungatei and Methanobacterium formicicum (H2 and formate-utilizing methanogens), Methanobrevibacter arboriphilus (only H2-utilizing methanogens) and Methanosaeta concillii (acetate-utilizing methanogen).
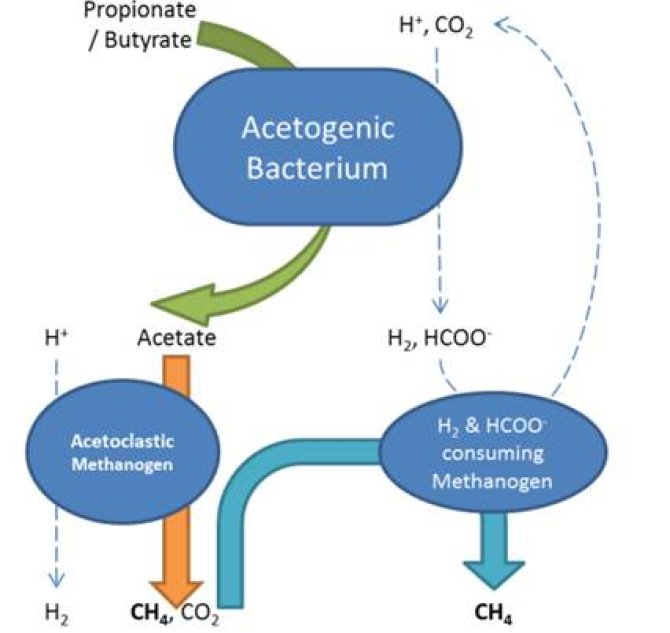
Defined cocultures will be grown in chemostat cultures and determine substrate consumption, product formation and biomass formation in response to changes in the environmental conditions. These kinetic data will be linked to the physiological features of the microorganisms involved. Presently, the fastest way to get insight into regulatory features of mixed communities is through differential proteomics. By using proteomic approaches at each growth condition the levels of key redox enzymes involved in interspecies electron transfer can be determined. This allows to make a systems biology approach to get insight into the metabolic interactions of syntrophic fatty acid-degrading communities.
If you have further questions or interests in the subject please contact me.