Project
Internship: Erasmus MC Rotterdam Identificationof Human papillomavirus variants (HPV) in cervical cancer and determination ofclinical significance
Group: Pathology and Clinical Bioinformatics, Erasmus MC, Rotterdam (EMC)
https://www.erasmusmc.nl/en/research/departments/pathology
Supervisors: Andrew Stubbs (EMC), Arlene Oei (AMC) Peter van Baarlen (HMI WU)
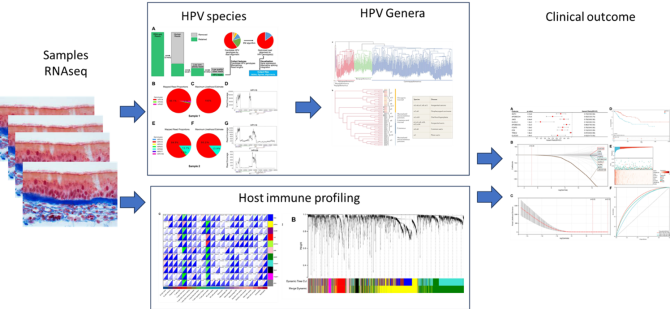
Background and research interests - Human Papilloma Virus (HPV) is a causative agent for the development and progression of multiple tumour types. HPV subtype has been demonstrated to be associated either favourably or unfavourably with clinical outcome. Thus, accurate HPV genotyping is crucial in facilitating epidemiology studies, vaccine trials, and HPV-related cancer research. Contemporary HPV genotyping assays only detect < 25% of all known HPV genotypes and are not accurate for low-risk or mixed HPV genotypes. Current genomic HPV genotyping algorithms use a simple read-alignment and filtering strategy that has difficulty handling repeats and homology sequences whilst novel methods are capable of circumventing these challenges. The immune profile of HPV patients has been corrected to survival (Wei et al, 2023; https://doi.org/10.3389/fonc.2022.860900) and we wish to extend these findings to the cohort of patients obtained in collaborations with Dr A.L. Oei (AMC)
Objectives
The aims of the study are to:
- Accurately determine the HPV species, immune phenotype and expression levels in each biopsy;
- Correlate these findings with clinical outcome
Methodology / what students can learn
The student will implement and validate existing HPV detection workflows and extend these where necessary to fulfil the scope of the project. The student will be required to determine the immune profiles and marker genes associated with HPV and relate this to clinical outcome. If time permits, these results might be translated into a risk model.
The student will learn to and be expected to develop, execute, manage, report and complete the research plan developed in collaboration with EMC and AMC.
Requirements
We are looking for Bioinformatics MSc students interested in developing their analytical research skills more advanced analytical techniques. The student should be proficient in python and R. The project will have a duration of 6 months.
Contact information
Andrew Stubbs, Pathology and Clinical Bioinformatics – a.stubbs@erasmusmc.nl
Arlene L. Oei, Lab. for Exp. Oncology and Radiobiology (LEXOR) / Department of Radiation Oncology - a.l.oei@amsterdamumc.nl
Peter van Baarlen, Host-Microbe Interactomics, Wageningen University, Animal Sciences - peter.vanbaarlen@wur.nl