Project
The side-effects of fisheries induced bottlenecks: Gauging extinction risk from the genome of blacktip reef sharks
This study will serve as a model for the application of genomics in the conservation biology in marine ecosystems. We will provide a profound resource for understanding how wild marine predators respond to the negative anthropogenic effects of overfishing and climate change; a concern facing many fisheries on a global scale.
Motivation
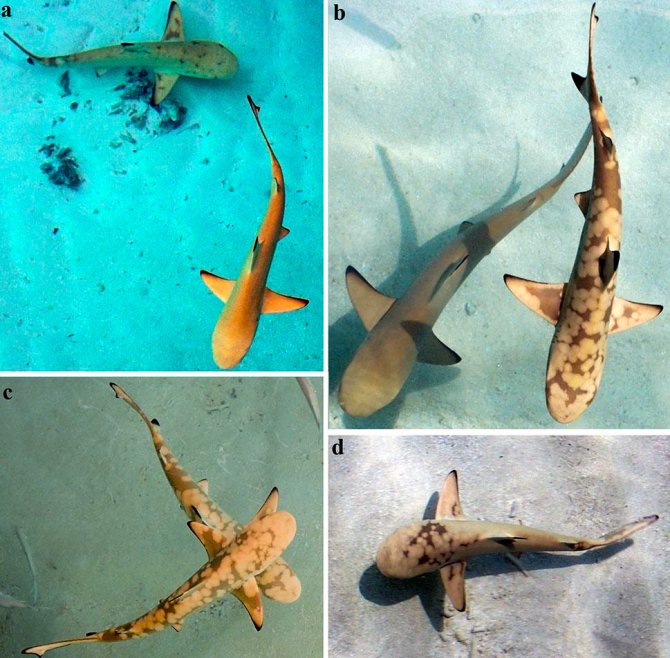
We will design functional markers for inbreeding risk that can be applied by sampling water from the reef, in the form of species-specific eDNA analysis. Such analyses can provide routine, cheap, completely non-invasive assessment of critical population parameters (e.g. locala effective population size) and occurrence of deleterious variation for species monitoring.
Aim of objections
- Infer the recent inbreeding history of the blacktip reef shark (Carcharhinus melanopterus) throughout the Maldivian archipelago.
- Determine the presence of functional detrimental variants and connect them to the phenotypic data obtained from observations in the wild.
- Characterize the functional genomic basis for inbreeding risk, specifically directed towards deleterious alleles that impact the survival of juveniles.
- To create non-invasive tools to monitor inbreeding risk in wild populations of C. melanopterus.
Method
Traditionally, species and populations monitoring requires exhaustive sampling and tracking over extended periods of time. These sampling and tracking techniques usually require species and habitat specific approaches that are time consuming and expensive, for marine species in particular. Conversely, recent advancements in genomics technology has allowed for functional and population genomics insights with a fraction of the individuals required for species and populations monitoring. To further understand a skin disorder in the blacktip reef shark, the disease phenotype will be characterized through photographs and acoustic tracking data during field-based surveys. Fine-scaled movements through acoustic tracking in combination with genomics will allow for elucidation of connectivity between reef shark populations (e.g. gene flow and population differentiation). Ultimately, long-term monitoring will be the primary vector for conservation management. We will use environmental DNA (eDNA) to non-invasively track the presence of deleterious alleles.