Project
The influence of proteins on the large-scale configurations of DNA
DNA is finding more and more non-biological applications as a highly versatile material for building molecular structures, such as in DNA origami (building 3D nanostructures) and DNA templating (for example metallizing DNA to make nanowires).

For these non-biological applications, but also for many biotechnological and medical applications, one can image that it is useful to be able to change DNA properties, for example using polymers/proteins that bind to the DNA. We are designing and producing artifical polymer-like proteins that coat single DNA molecules in a similar way as viral capsid proteins do. The properties of the complexes of these protein polymers with DNA are studied using techniques such Dynamic Light scattering, Atomic force Microscopy, Surface Plasmon Resonance and Fluorescence Correlation Spectroscopy.
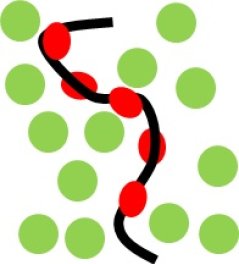
DNA condensation by binding and non-binding proteins
The large-scale configurations of DNA in living cells are shaped to a large extent by proteins, both DNA binding proteins and non-DNA binding proteins. Binding proteins may wrap, bend and cross-link DNA, whereas non-binding proteins may push DNA into condensed states. Using simulations, simple analytical theories and experiments on model systems, I try to understand the basics of how proteins influence the large-scale configurations of DNA.
Self Assembled Protein Based Block Copolymers for Encapsulation and Delivery of DNA
We work designing protein block copolymers for co-assembly with DNA and negative charged polyelectrolytes. The protein block copolymers are prepared using DNA recombinant technology and produced through fermentation of recombinant microorganisms. We combine several functional blocks such as Collagen-like, Silk-like and others that fold and self assemble into predictable secondary and supramolecular structures. The main objective is to design block copolymers able to co- assemble upon interaction with DNA in a cooperatively and stable way such as viral coat proteins do. Besides the design work, we are looking to understand the physical chemistry principles that govern such behavior in order to tune the properties. Between the applications we are interested to test the designs include gene therapy and drug delivery, DNA nanotechnology, bioelectronics, etc.
We use techniques such as Molecular Biology for production (FBR-WUR). The characterization is carried out using Light scattering techniques, atomic force microscopy, TIRF, fluorescence microscopy, spectroscopic methods, surface plasmon resonance, between others.
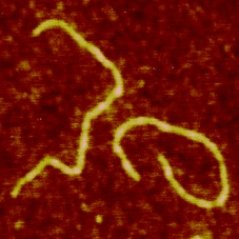
Our current results include development of a family of diblock copolymers able to coat single DNA molecules into semi flexible rods (Fig 1). Currently we have produced a triblock carrying a cooperative block where self assembly is triggered by electrostatic interaction with DNA (Fig. 2).
The general project includes collaboration with an Eindhoven team that makes computer simulations about folding of protein secondary structures such as beta sheet strands with cationic corners triggered by electrostatic interaction of DNA.
Funding: DPI - CRP project 698 "Designer Polypeptides for Self-Assembled Delivery Vehicles"Collaboration with Paul van der Schoot, Tue
More information
See: How to Make an Artificial Virus? : Decodifying the Viral Coat Protein
de Vries, R"DNA condensation in bacteria: interplay between macromolecular crowding and nucleoid proteins"
Biochimie 92, 1715-1721 (2010)
http://dx.doi.org/10.1016/j.biochi.2010.06.024
Ramos Bessa, J.E.B..;de Vries, R."Polymer induced condensation of DNA supercoils"
The Journal of Chemical Physics 129, 185102 (2008)
http://dx.doi.org/10.1063/1.2998521
Ramos, E.B.;Wintraecken, K.;Geerling, A.;de Vries, R."Synergy of DNA-bending nucleoid proteins and macromolecular crowding in condensing DNA"
Biophysical Reviews and Letters 2, 259-265 (2007)
http://dx.doi.org/10.1142/S1793048007000556
de Vries, R."Depletion-induced instability in protein-DNA mixtures: Influence of protein charge and size"
The Journal of Chemical Physics 125, 014905 (2006) http://dx.doi.org/ doi:10.1063/1.2209683
de Vries, R."Evaluating changes of writhe in computer simulations of supercoiled DNA"
The Journal of Chemical Physics 122, 064905 (2005) http://dx.doi.org/10.1063/1.1846052
Ramos Bessa, J.;de Vries, R.;Ruggiero Neto, J."DNA Ψ-Condensation and Reentrant Decondensation: Effect of the PEG Degree of Polymerization"
The Journal of Physical Chemistry B 109, 23661-23665 (2005) http://dx.doi.org/10.1021/jp0527103=
de Vries, R."Flexible Polymer-Induced Condensation and Bundle Formation of DNA and F-Actin Filaments"
Biophysical Journal 80, 1186-1194 (2001) http://dx.doi.org/10.1016/S0006-3495(01)76095-X