Genetic selection against endemic infections in cattle: is eradication of Mycobacterium avium subspecies paratuberculosis possible?
WIAS Magazine - Fall edition 2023
Research Overview
Keywords: MAP, disease modeling, genetic selection, eradication, intervention
Background
Paratuberculosis (Johne’s disease) is a chronic gastrointestinal disease of cattle caused by Mycobacterium avium subsp. paratuberculosis (MAP) and is now known to be endemic worldwide (Johne & Frothingham, 1895). The disease is of economic interest to dairy farms due to reduced milk production, early culling, and increased mortality (Bush, Windsor, & Toribio, 2006), and is of public health interest to humans due to its potential association with Crohn’s disease (Frank, 2008; Shafran, 2008). Control programmes have been implemented in several countries, but eradication is still a challenge. The difficulties in eradicating Paratuberculosis lie in the technical limitations of diagnosis (Collins et al., 2006), the lack of practical treatments (Harris & Barletta, 2001), the ineffective vaccine and the complexity and difficulty of the prevention and control measures at the farm level (Geraghty et al., 2014). Thus, attention was turned to investigate the feasibility of eradication of Paratuberculosis by using genetic selection as an additional intervention strategy. This approach seems reasonable because cattle resistance to Paratuberculosis has been shown to be heritable (Attalla et al., 2010; McGovern et al., 2019; Mortensen, Nielsen, & Berg, 2004; van Hulzen et al., 2011). Long-term data on MAP are available for all (dairy) cattle in Netherlands. Therefore, we may be able to select for MAP resistance by using phenotypic and genomic data. Also, although individuals are infected for life, infection occurs mainly early in life, so we can observe the effects of intervention relatively quickly (Windsor & Whittington, 2010).
Objectives
- Modelling of MAP transmission within Dutch dairy herds, identifying key parameters that can be used as target for genetic interventions
- Estimating genetic variation and heritability related to MAP susceptibility/ infectivity for those parameters in Dutch Holstein cattle
- Performing genomic prediction and genome-wide association studies (GWAS) to estimate breeding value and to identify genomic regions and candidate genes associated with preferred traits
- Scenario study for validation of interventions
- Developing a breeding strategy to get rid of MAP
Approach:
Firstly, we will collect phenotype and genotype data from external parties (CRV and GD). Secondly, we will develop a stochastic continuous-time, age-structured, environmental transmission model to simulate the accumulation as well as the decay of MAP that are shed into the environment (Fig. 1).
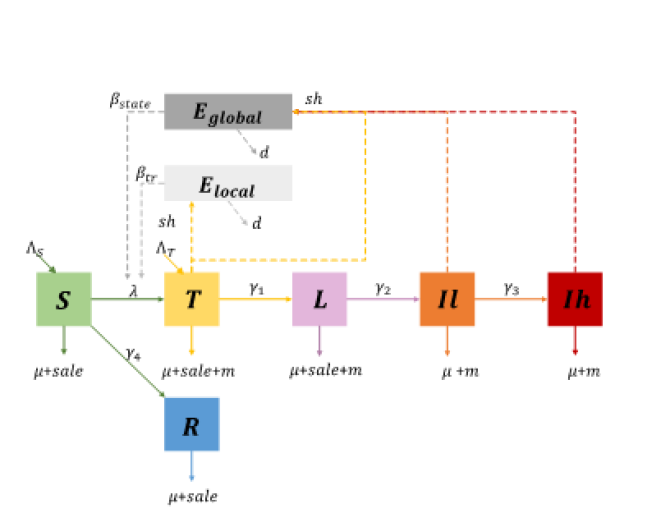
Figure 1. Transmission model for MAP in a Dutch dairy herd. Health states in the specific age- structured (a) population include susceptible (S), resistant (R), transiently infectious (T), latently infected (L), lowly infectious (Il), highly infectious (Ih). The amount of MAP in the collective pen is represented as the hazard level of local environment (Elocal), and the amount of MAP in the farm is represented as the hazard level of the global environment (Eglobal). The birth rate of a susceptible calf or an infected calf (congenital or infected in the first week) is ^S, ^T respectively. The infection rate in the environmental transmission model is λ and the transmission rate is β. The transition rates between each compartment are γ1, γ2, γ3 and γ4. The natural mortality rate is age-dependent, denoted as μ. The mortality rate caused by disease is dependent on infected status, denoted as m. The sale rate of the male calf is expressed as sale.
Building on this model, the following step is to carry out a genetic analysis to estimate heritability of Paratuberculosis susceptibility and infectivity in Dutch dairy cattle and to estimate breeding values. A general linear model (GLM), which has been developed by Biemans et al.(2017), will be used as a starting point and then adapted to the characteristics of MAP. Thirdly, GWAS will be carried out to identify candidate genes associated with preferred traits and their functional annotation. Finally, the goal is to discuss the feasibility and propose a breeding strategy. A scenario study will be applied to quantify the contribution of genetic selection, as well as other interventions, to the eradication of MAP (Fig. 2).
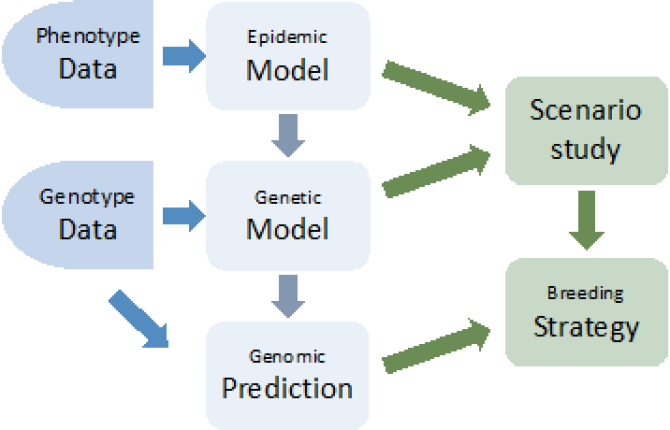
Figure 2. Proposal project flowchart.
Perspectives
Since existing prevention and control measures have not led to eradication of MAP, it’s innovative to explore the feasibility of genetic selection as an additional measure to achieve that goal. The inclusion of genetic effects into epidemiological models is a novel and promising development. With this project, we want to further develop knowledge and tools to better understand the transmission of MAP in dairy farms and enable breeders to effectively select against MAP.
References
Attalla, S. A., Seykora, A. J., Cole, J. B., & Heins, B. J. (2010). Genetic parameters of milk ELISA scores for Johne's disease. Journal of Dairy Science, 93(4), 1729-1735. https://doi.org/https://doi.org/10.3168/jds.2009-2625
Biemans, F., de Jong, M. C. M., & Bijma, P. (2017). A model to estimate effects of SNPs on host susceptibility and infectivity for an endemic infectious disease. Genetics Selection Evolution, 49(1), 53. https://doi.org/10.1186/s12711-017-0327-0
Bush, R., Windsor, P., & Toribio, J. A. (2006). Losses of adult sheep due to ovine Johne's disease in 12 infected flocks over a 3‐year period. Australian Veterinary Journal, 84(7), 246-253.
Frank, D. N. (2008). Mycobacterium avium subspecies paratuberculosis and Crohn's disease. The Lancet infectious diseases, 8(6), 345.
Geraghty, T., Graham, D. A., Mullowney, P., & More, S. J. (2014). A review of bovine Johne's disease control activities in 6 endemically infected countries. Preventive Veterinary Medicine, 116(1), 1-11. https://doi.org/https://doi.org/10.1016/j.prevetmed.2014.06.003
Harris, N. B., & Barletta, R. G. (2001). Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clinical microbiology reviews, 14(3), 489-512.
Johne, H., & Frothingham, L. (1895). Ein eigenthuemlicher fall von tuberculose beim rind. Dtsch. Z. Tiermed. Pathol, 21, 438-454.
McGovern, S. P., Purfield, D. C., Ring, S. C., Carthy, T. R., Graham, D. A., & Berry, D. P. (2019). Candidate genes associated with the heritable humoral response to Mycobacterium avium ssp. paratuberculosis in dairy cows have factors in common with gastrointestinal diseases in humans. Journal of Dairy Science, 102(5), 4249-4263. https://doi.org/https://doi.org/10.3168/jds.2018-15906
Mortensen, H., Nielsen, S. S., & Berg, P. (2004). Genetic Variation and Heritability of the Antibody Response to Mycobacterium avium subspecies paratuberculosis in Danish Holstein Cows. Journal of Dairy Science, 87(7), 2108-2113. https://doi.org/https://doi.org/10.3168/jds.S0022-0302(04)70029-6
Shafran, I. (2008). Potential Pathogenic Role of Mycobacterium avium Subspecies paratuberculosis in Crohn's. Inflamm Bowel Dis, 14(12), 1753.
van Hulzen, K. J. E., Nielen, M., Koets, A. P., de Jong, G., van Arendonk, J. A. M., & Heuven, H. C. M. (2011). Effect of herd prevalence on heritability estimates of antibody response to Mycobacterium avium subspecies paratuberculosis. Journal of Dairy Science, 94(2), 992-997. https://doi.org/https://doi.org/10.3168/jds.2010-3472
Windsor, P. A., & Whittington, R. J. (2010). Evidence for age susceptibility of cattle to Johne’s disease. The Veterinary Journal, 184(1), 37-44. https://doi.org/https://doi.org/10.1016/j.tvjl.2009.01.007