News
S. suis in tonsil-associated microbiome of piglets
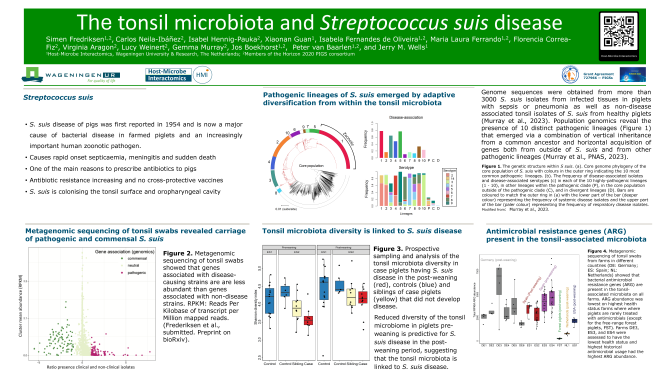
Metagenomic sequencing of the tonsil-associated microbiome of piglets revealed that both pathogenic and commensal Streptococcus suis are carried in piglets on many farms. “This poses a risk for further evolution of the bacteria”, says Professor Jerry Wells of the chair group Host-Microbe Interactomics of Wageningen University & Research. The results of the research were presented on a poster in connection with the University Dies Natalis.
Research
Streptococcus suis disease in piglets was first reported in 1954 and has become an increasing problem in the pig rearing industry worldwide. “S. suis disease typically manifests as sepsis and rapid onset meningitis, making it a serious welfare issue”, explains Professor Jerry Wells of the HMI chair group. There are no cross-protective vaccines on the market and antibiotics are commonly used to control spread of S. suis in post weaning piglets, contributing to the spread of antimicrobial resistance. S. suis is also an emerging human pathogen with highest cases in Southeast Asia.
Population genomics
“Most cases of S. suis disease globally are caused by a small number of highly pathogenic lineages”, states Wells. This observation was presented at the Microbiome event and reported on a recent population genomics study by Murray et al. (Proc Natl Acad Sci U S A. 2023 Nov 21;120 (47)) and involving WUR partners in the Horizon 2020 PIGSs project. “The pathogenic strains emerged from a sub-population of diverse and largely commensal S. suis species also residing on the tonsil and upper respiratory tract, through acquisition of specific genomic pathogenicity-associated islands.”
Gene transfer
Using metagenomic sequencing Simen Fredriksen and his colleagues of the chair group HMI (WUR) showed that the tonsil-associated microbiome of piglets harbors both commensal and pathogenic S. suis which tend to have smaller genomes and fewer genes. These results show the potential for continued evolution and diversification of pathogenic S. suis through intra- and inter-species horizontal gene transfer with the resident microbiota. “This poses a risk for evolution of pathogenic S. suis that can efficiently colonise human hosts, leading to increased infection of humans”, explains Wells.
Diversity decrease
This study also showed that decreased diversity of the tonsil microbiota of piglets was linked to S. suis disease post-weaning. Furthermore the tonsil-associated microbiota on all European farms studied carried numerous antibiotic resistance genes (ARGs). “ARG abundance was lowest on highest health status farms where piglets are rarely treated with antimicrobials, confirming that the failure to prevent bacterial infections leads to increased AMR”, concludes Wells.